Tracing Ancestry, Researchers Produce a Genetic Atlas of Human Mixing Events, by Nicholas Wade, New York Times, 13 Feb 2014:
Having sampled genomes from around the world, they found they could detect about 95 distinguishable populations.
Though all humans have the same set of genes, their genomes are studded with mutations, which are differences in the sequence of DNA units in the genome. These mutations occur in patterns because whole sets of mutations are passed down from parent to child and hence will be common in a particular population. Based on these patterns, geneticists can scan a person’s genome and assign the ancestry of each segment to a particular race or population.
The Myers group has posted its results on a web page that records the degree of admixture in each population. The English, however, known to be a rich medley of Celts with invaders such as the Angles, Saxons, Jutes, Danes and Norwegians, carry the notation “No strong evidence of admixture.” Dr. Myers said his method cannot yet detect genetic mixing between very similar populations, as was the case with the English and their invaders from Scandinavia and Northern Germany. He said he hoped to distinguish all these groups in a separate project on British ancestry.
The semitically correct emphasis is on admixture/mixing, though a closer look reveals the reality of race.
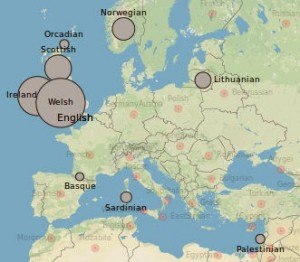
A genetic atlas of human admixture history, “Companion website for “A genetic atlas of human admixture history”, Hellenthal et al, Science (2014)”, lists the 95 “target populations” they distinguished:
Adygei, Armenian, Balochi, BantuKenya, BantuSouthAfrica, Basque, Bedouin, Belorussian, BiakaPygmy, Brahui, Bulgarian, Burusho, Cambodian, Chuvash, Colombian, Cypriot, Dai, Daur, Druze, EastSicilian, Egyptian, English, Ethiopian, EthiopianJew, Finnish, French, Georgian, GermanyAustria, Greek, Hadza, Han, HanNchina, Hazara, Hezhen, Hungarian, Indian, IndianJew, Iranian, Ireland, Japanese, Jordanian, Kalash, Karitiana, Lahu, Lezgin, Lithuanian, Makrani, Mandenka, Maya, MbutiPygmy, Melanesian, Miao, Mongola, Moroccan, Mozabite, Myanmar, Naxi, NorthItalian, Norwegian, Orcadian, Oroqen, Palestinian, Papuan, Pathan, Pima, Polish, Romanian, Russian, Sandawe, SanKhomani, SanNamibia, Sardinian, Saudi, Scottish, She, Sindhi, SouthItalian, Spanish, Surui, Syrian, Tu, Tujia, Tunisian, Turkish, Tuscan, UAE, Uygur, Uzbekistani, Welsh, WestSicilian, Xibo, Yakut, Yemeni, Yi, Yoruba
“Population genetics” research like this is a form of race science, with “population” serving as a rough euphemism for “race”, because in a broad sense a race is a cluster of relatively closely-related people. The clusters at the coarsest, continental scale are the ones most commonly associated with the word race today – the White, black, yellow and red races. But at the ethnic/national scale there are more clusters, like the 95 groups identified by this atlas. An even closer examination of the DNA would tease apart even finer distinctions, resolving larger clusters into sub-ethnic, clan and family strands, right on down to positively identifying an individual’s parents or siblings.
The propaganda that race is a social construct based on superficial attributes such as skin color, is a big lie. Race and racial distinctions are deeply rooted in biology. Though there are many clusters, at different scales, and the boundaries of those clusters are fuzzy, they are not arbitrary. As the article notes, “geneticists can scan a person’s genome and assign the ancestry of each segment to a particular race or population”. The same clusters people can distinguish with their senses alone correspond to the distinctions determinable by DNA alone.
Wade’s remark about the English reflects the clash between the jewish “nation of immigrants” narrative and the reality of Northwestern European genetic homogeniety.
Curiously, the atlas includes EthiopianJew and IndianJew but not the more numerous Ashkenazi and Sephardic clusters. Wade reported on the clustering of jews in June 2010, Studies Show Jews’ Genetic Similarity:
The shared genetic elements suggest that members of any Jewish community are related to one another as closely as are fourth or fifth cousins in a large population, which is about 10 times higher than the relationship between two people chosen at random off the streets of New York City, Dr. Atzmon said.
Ashkenazic and Sephardic Jews have roughly 30 percent European ancestry, with most of the rest from the Middle East, the two surveys find. The two communities seem very similar to each other genetically, which is unexpected because they have been separated for so long.
The research this NYT report discusses, Abraham’s Children in the Genome Era: Major Jewish Diaspora Populations Comprise Distinct Genetic Clusters with Shared Middle Eastern Ancestry, concludes:
This study touches upon an issue that was raised over a century ago by Maurice Fishberg, Joseph Jacobs, and others about whether the Jews constitute a race, a religious group, or something else.29,30 In this study, Jewish populations from the major Jewish Diaspora groups—Ashkenazi, Sephardic, and Mizrahi—formed a distinctive population cluster by PCA analysis, albeit one that is closely related to European and Middle Eastern, non-Jewish populations. Within the study, each of the Jewish populations formed its own cluster as part of the larger Jewish cluster. Each group demonstrated Middle Eastern ancestry and variable admixture with European populations.
Whatever you want to call them, the point is that jews can be distinguished by their DNA. In fact, though neither Wade nor Hellenthal et al mention any jews in the “rich medley” of England, it’s not because finer analysis is required to detect them there, or anywhere else in Northwestern Europe. Finding “Lithuanian” and “Palestinian” admixture in the English, as illustrated by the attached image, seems to be a way of detecting the jews without noticing them.